Contents
Nucleic acid analogues
Peptide nucleic acid (PNA)
Peptide nucleic acid (PNA) is a nucleic acid analogue in which the normal phosphate linkage found in DNA and RNA is replaced by an neutral peptide-like N-(2-aminoethyl)glycine backbone (Figure 1, Figure 2).
Synthesis
PNA synthesis is usually performed by automated solid-phase methods, as with DNA and RNA oligonucleotide synthesis. The mechanism of PNA synthesis has much in common with solid-phase peptide synthesis.
Solid-phase peptide synthesis
The coupling step in peptide synthesis is an amide bond coupling reaction between two amino acids (Figure 3). The growing peptide chain is attached to the resin via an ester or amide linkage, and the amine of the new unit is protected with a 9-fluorenylmethyloxycarbonyl (Fmoc) protecting group.
The carboxylic acid must be activated in order for the coupling reaction to proceed. The carboxylic acid is first converted to a O-acylisourea by reaction with a carbodiimide. Formation of diisipropylurea drives the reaction to completion. The O-acylisourea, which is susceptible to rearrangement to the unreactive N-acylurea, is spontaneously converted to a reactive 1-hydroxybenzotriazole ester, by reaction with 1-hydroxybenzotriazole.}
Solid-phase PNA synthesis
As in solid-phase peptide synthesis, the coupling step in PNA oligonucleotide synthesis is an amide bond coupling reaction between two amino acids (Figure 5).
In PNA synthesis, the carboxylic acid is activated using a benzotriazole uronium salt, such as O-(1H-benzotriazol-1-yl)-N,N,N',N'-tetramethyluronium hexafluorophosphate (HBTU), which, like the combination of HOBt and a diimide, yields a hydroxybenzotriazole ester. The driving force for the reaction is the formation of the by-product tetramethylurea (Figure 6).
The Fmoc protecting groups are removed by treatment of the PNA oligonucleotide with a solution of 20% piperidine in DMF (Figure 7) at the end of each PNA synthesis cycle.
Structures of PNA monomers
As the chemistry of PNA oligonucleotide synthesis is different from that of DNA and RNA oligo synthesis, different types of protecting groups are needed for the heterocyclic bases. The benzhydryloxycarbonyl (Bhoc) protecting group is used to protect the primary amines of the heterocyclic bases in PNA oligo synthesis. (Figure 8).
Synthesis of PNA monomers, and resin functionalization
The Bhoc-protected heterocyclic base (or unprotected thymine) is alkylated at the N(2) position with bromoacetic acid. The resulting nucleobase acetic acid is then coupled with an Fmoc-protected 2-((2-aminoethyl)amino) acetate using a coupling reagent (e.g. a carbodiimide), to yield the monomer (Figure 9a). Functionalization of the resin with the PNA monomer is a simple matter of coupling the monomer with the free resin (Figure 9b).
Deprotection of heterocyclic bases
The Bhoc protecting groups on the heterocyclic bases, and the linker between the PNA and the solid support, are acid-labile, and so can be removed in a single step by treatment with trifluoroacetic acid at the end of PNA assembly (Figure 10).
Properties and uses
PNA oligonucleotides have unique properties that have been utilized in many molecular biology and biochemical applications.
PNA oligonucleotides hybridize with complementary DNA and RNA oligonucleotides, by Watson-Crick base pairing. Perhaps surprisingly, PNA oligonucleotides hybridize to complementary DNA/RNA with higher affinity and specificity than normal DNA/RNA oligonucleotides. PNA is not a nucleic acid, but a neutral molecule. The lack of a negatively charged backbone reduces electrostatic repulsion between oligonucleotides in PNA:DNA and PNA:RNA hybrids, contributing to increased stability. In contrast to DNA:DNA duplexes, the stability of PNA:DNA duplexes is practically independent of salt concentration.
PNA probes
Like DNA and RNA, PNA oligonucleotides can be labelled with fluorescent dyes during or after solid-phase synthesis. These PNA probes are used in diagnostics and detection.
The increased specificity of PNA oligonucleotides in forming hybrid duplexes with DNA and RNA means that mismatched base pairs in hydrid PNA:DNA duplexes are particularly unstable. PNA probes are therefore useful in identifying point mutations and SNPs. The destabilizing effect of the mismatch can be increased by the use of very short PNA probes. DNA probes often fail to distinguish between certain regions of highly structured RNA, but PNA probes can be used to detect these regions, if the hybridization is carried out at low salt concentration so that the RNA is unstructured.
PNA probes are also used in fluorescent in situ hybridization (PNA-FISH). In fluorescent in situ hybridization (FISH), short probes can be difficult to detect (owing to insufficient hybridization and low levels of labelling), while longer probes can induce non-specific background fluorescence. In PNA-FISH, shorter PNA probes can be used (because of the higher specificity of PNA), resulting in less background noise.
Because of their insensitivity to salt concentration, PNA probes can often be used in a wider range of conditions than DNA and RNA probes, which can be useful in applications such as microarrays.
Therapeutic applications
PNA is an unnatural molecule, and PNA oligonucletides are not substrates for enzymes that modify DNA, RNA and proteins. DNA and RNA oligonucleotides are effective in antisense therapy in vitro, but are degraded quickly by intracellular nucleases in vivo, and this degradation destroys their antisense activity. PNA oligomers are resistant to degradation by nucleases and are therefore much more effective than DNA and RNA in therapeutic antisense gene silencing.
PNA in molecular biology
The lack of reactivity of PNA oligonucleotides to nucleic acid- and protein modifying enzymes means that PNA oligonucleotides are incompatible with useful molecular biology techniques such as PCR, restriction enzymes and proteolysis. However, this allows PNA to used along with DNA in the presence of enzymes: while the DNA is modified by the enzyme, the PNA remains intact. PNA oligonucleotides can be used in the presence of nucleases to form an "artificial restriction" system, enabling the cutting of DNA at sites not accessible using restriction enzymes. PNA oligonucleotides can also be used in PCR, enhancing the amplification of specific DNA products.
Locked nucleic acid (LNA)
Locked nucleic acid (LNA) is an unnatural conformationally restricted oligonucleotide analogue bearing a close structural resemblance to DNA, but possessing monomer units with a 2'-O-4'-C-methylene bridge. The bicyclic structure locks the molecule in a C3'-endo sugar (N-type) configuration (Figure 11), ensuring the oligonucleotide adopts the A-form helix (which is associated with high duplex stability).
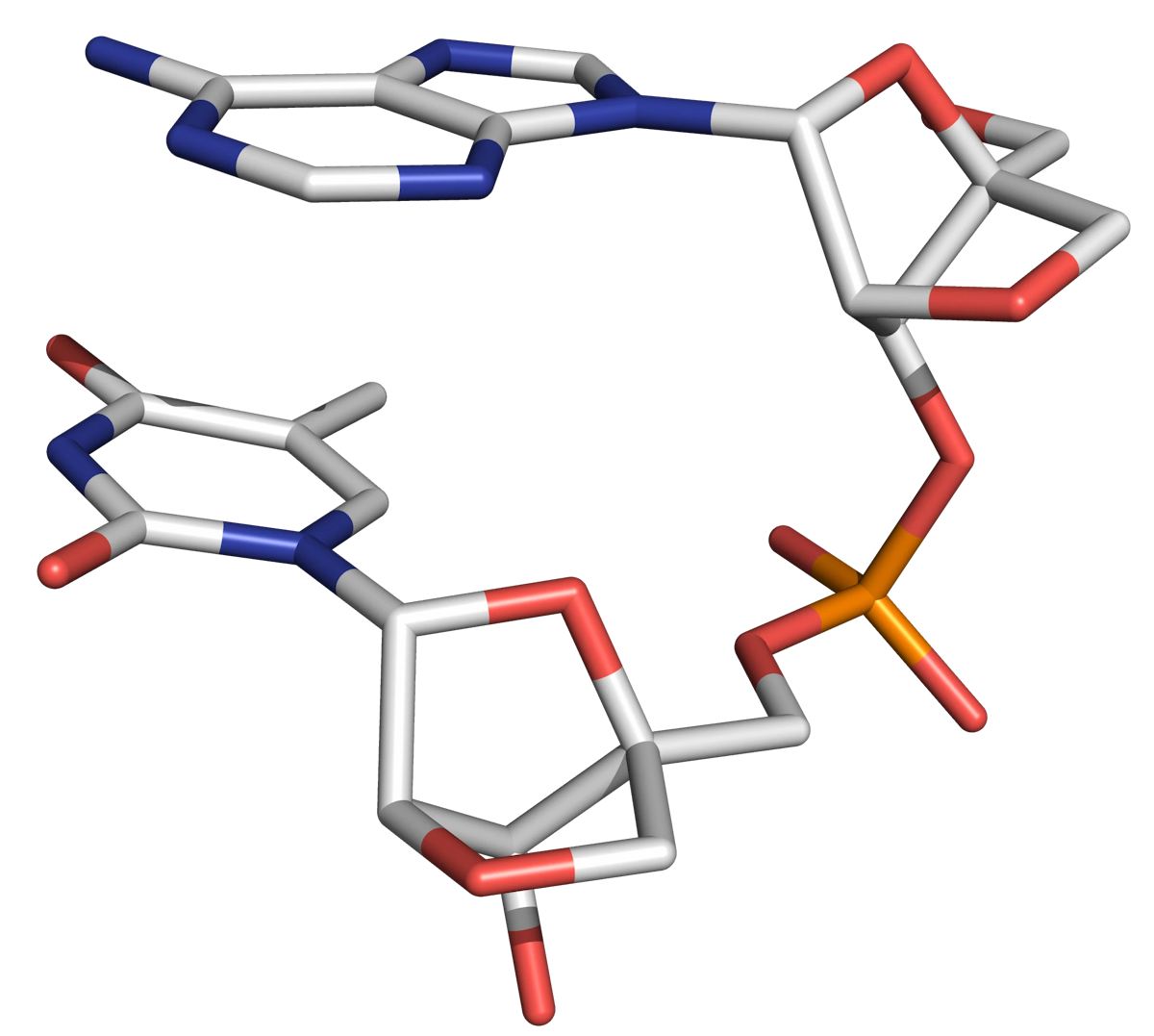
The original LNA oligomers were "oxy-LNAs". Subsequently "thio-LNA" (2'-S-CH2-4') and "amino-LNA" (2'-NH-CH2-4') analogues were prepared (Figure 13).
Synthesis
LNA oligonucleotides (and mixed LNA/DNA and LNA/RNA oligomers) can be synthesized on standard automated DNA synthesizers using commerically available LNA phosphoramidite monomers (Figure 14). The only changes to the standard DNA synthesis cycle are increased reaction times for the coupling and oxidation steps. LNA oligonucleotides can be labelled with fluorescent dyes and other chemical modifications in the same way as DNA and RNA oligonucleotides.
Properties and uses
LNA forms stable hybrid duplexes with DNA and RNA. Duplexes formed between two LNA oligonucleotides exhibit very high affinity and specificity. LNA units can be incorporated into normal DNA or RNA oligonucleotides to increase the stability of probe-target hybrids, manifested as an increased melting temperature (Tm) of the hybrid duplex. LNA oligonucleotides can be used for mismatch discrimination, their high binding affinity allowing short probes to be used. LNA oligos have been used in triplex forming oligonucleotides, antisense oligonucleotides and microarrays.
The incorporation of LNA bases into antisense DNA oligonucleotides provides both strong affinity towards the RNA target and stability to nucleases. One of these gapmers, EZN-3042, has proved effective in inhibiting tumour growth and is currently in clinical trials as a leukaemia drug.
Unlocked nucleic acid (UNA)
Unlocked nucleic acid (UNA) is an analogue of RNA in which the C2'-C3' bond has been cleaved. Whereas LNA is conformationally restricted (relative to DNA and RNA), UNA is very flexible (Figure 15).
Synthesis
UNA can be synthesized using an automated solid-phase approach, using UNA phosphoramidite monomers (Figure 16).
Properties and uses
While the incorporation of LNA units into DNA and RNA oligonucleotides increases the stability of duplexes, UNA has a destabilizing effect. Incorporation of UNA units into an oligonucleotide probe lowers the Tm of the probe-target duplex. With their complementary effect on probe-target stability, LNA and UNA units can therefore be used to fine-tune the thermodynamic properties of DNA and RNA probes.
UNA bases have been incorporated into siRNA oligonucleotides, used in RNA interference (RNAi). While remaining potent, these UNA-modified siRNA (UsiRNA) molecules were found to be resistant to degradation in serum, have reduced toxicity and reduced off-target effects.
Triazole-linked DNA
Recently, triazole-linked DNA oligonucleotides, in which one or more of the natural phosphate backbone linkages are replaced with triazole linkages, have been synthesized using click chemistry.
The presence of a triazole linker does not perturb the global structure of double-stranded DNA relative to that of natural DNA. Hydrogen bonding between bases either side of the triazole linkage is not disrupted, and the overall structure of the helix is preserved. Small local changes are observed at the site of the triazole, but one of the nitrogen atoms of the triazole appears to mimic the phosphate group, a hydrogen bond acceptor, diminishing the effect of these local structural perturbations (Figure 17).
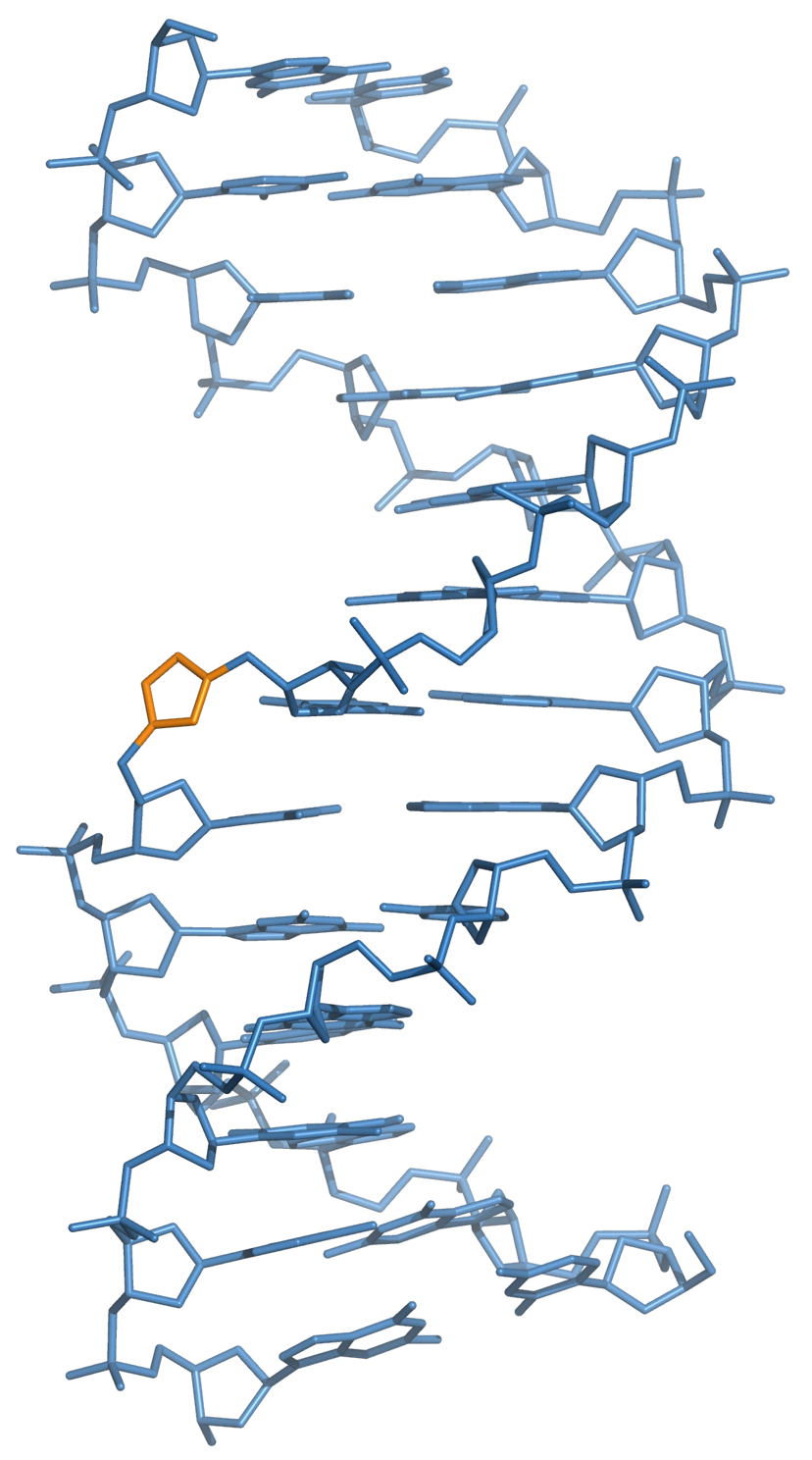
Synthesis by click chemistry
Triazole-linked oligonucleotides are assembled from smaller oligonucleotides with terminal alkyne and azide groups, in the copper-catalyzed azide-alkyne cycloaddition (CuAAC) reaction, the canonical click chemistry reaction.
Properties and uses
Triazole-linked oligonucleotides form duplexes with complementary strands of natural DNA. Triazole-linked DNA is read through by DNA and RNA polymerases, and is functional as a PCR template in E. coli. Triazole-linked RNA has been used in the synthesis of an ribozyme: despite the presence of a triazole linkage at the active site, the triazole-linked ribozyme was found to be catalytically active.
The assembly of large oligonucleotides by "click" ligation of smaller oligonucleotides has the potential to overcome the practical size limit of around 200 bases that applies to solid-phase phosphoramidite oligonucleotide synthesis. Long oligonucleotides can be made by enzymatic ligation of shorter oligos, but this has problems: it does not work well on a large scale and does not tolerate epigenetic modifications. The chemical ligation of shorter oligos using click chemistry overcomes these problems, and could be used in the total synthesis of very long strands of DNA including synthetic genes.